Which of the following processes correctly describes alternative RNA splicing? This question delves into the intricate world of RNA processing, where a single gene can give rise to multiple protein isoforms through a process known as alternative RNA splicing. Alternative RNA splicing is a fundamental mechanism that contributes to the vast diversity of proteins in living organisms.
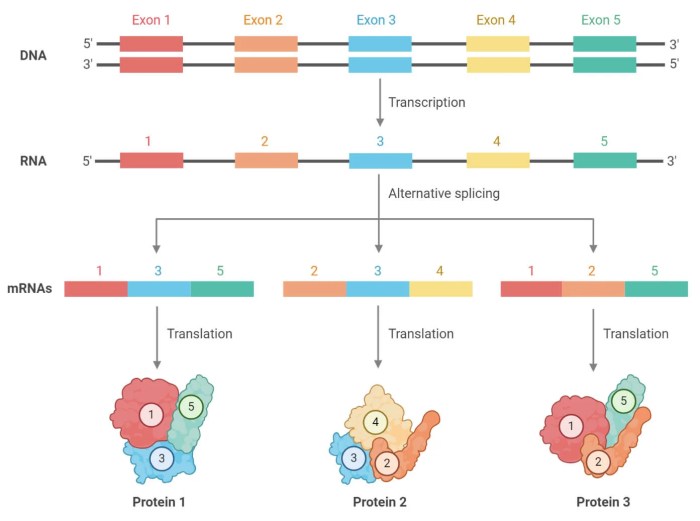
Alternative RNA splicing occurs when different combinations of exons and introns are spliced together to create different mature RNA molecules. This process is regulated by a complex interplay of splicing factors and RNA sequences, allowing cells to fine-tune gene expression and respond to changing cellular conditions.
Alternative RNA Splicing Overview
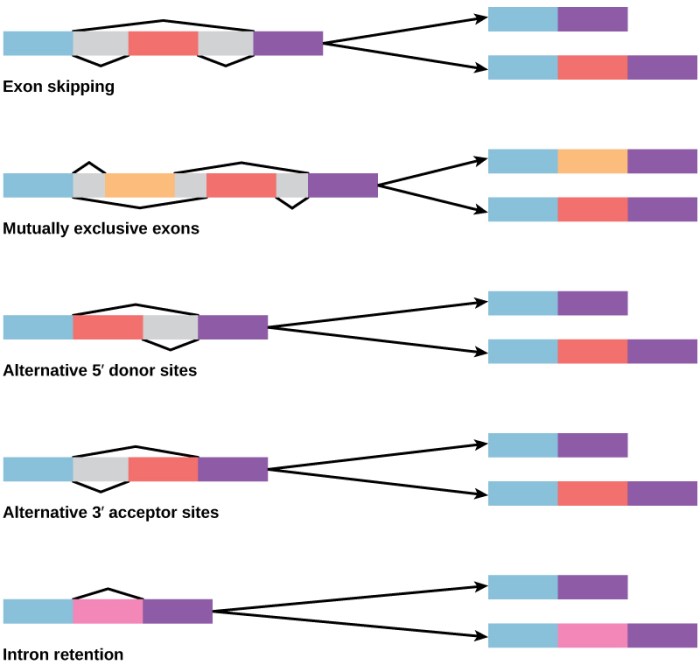
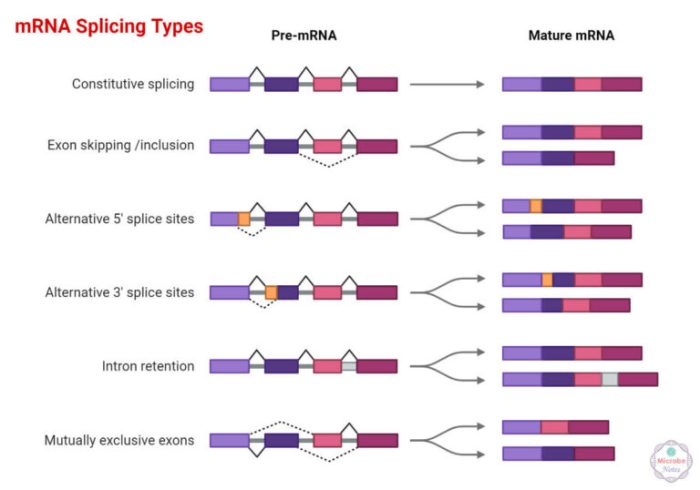
Alternative RNA splicing is a fundamental process that allows a single gene to encode multiple proteins. This process involves the selection of different splice sites within a pre-mRNA molecule, resulting in the production of different mature mRNA transcripts. These transcripts can then be translated into different protein isoforms, each with its unique structure and function.
There are several different types of alternative RNA splicing, including:
- Cassette Exon:An entire exon is either included or excluded in the mature mRNA.
- Intron Retention:An intron is retained in the mature mRNA instead of being spliced out.
- Alternative 5′ Splice Site:The splice site at the 5′ end of an exon is varied, resulting in different lengths of the 5′ untranslated region (UTR).
- Alternative 3′ Splice Site:The splice site at the 3′ end of an exon is varied, resulting in different lengths of the 3′ UTR.
Alternative RNA splicing is a highly regulated process that is essential for the proper function of eukaryotic cells. Defects in alternative RNA splicing can lead to a variety of diseases, including cancer, neurodegenerative disorders, and developmental abnormalities.
Mechanisms of Alternative RNA Splicing
Alternative RNA splicing is carried out by a large protein complex called the spliceosome. The spliceosome recognizes specific sequences within the pre-mRNA molecule and assembles at the splice sites. The spliceosome then catalyzes the splicing reaction, which involves the removal of the intron and the ligation of the exons.
The selection of splice sites is influenced by a variety of factors, including:
- Strength of the splice site:The strength of a splice site is determined by the sequence of nucleotides at the splice site. Strong splice sites are more likely to be used than weak splice sites.
- Presence of regulatory elements:Regulatory elements, such as silencers and enhancers, can influence the selection of splice sites. Silencers inhibit splicing, while enhancers promote splicing.
- Protein-protein interactions:Proteins can interact with the spliceosome and influence the selection of splice sites. These proteins can either promote or inhibit splicing.
Alternative RNA splicing is a highly regulated process that is essential for the proper function of eukaryotic cells. The regulation of alternative RNA splicing is complex and involves a variety of mechanisms, including transcriptional regulation, post-transcriptional regulation, and epigenetic regulation.
Consequences of Alternative RNA Splicing
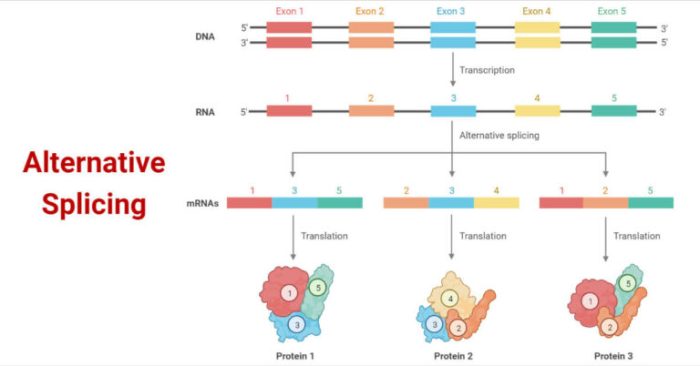
Alternative RNA splicing has a profound impact on protein diversity. A single gene can encode multiple proteins, each with its unique structure and function. This allows for a greater diversity of proteins to be produced from a limited number of genes.
Alternative RNA splicing also plays a role in gene regulation. The inclusion or exclusion of certain exons can alter the expression of a gene. For example, the inclusion of an exon that contains a stop codon can lead to the premature termination of translation.
This can result in the production of a truncated protein that is non-functional.
Defects in alternative RNA splicing can lead to a variety of diseases, including cancer, neurodegenerative disorders, and developmental abnormalities. For example, the mis-splicing of the BRCA1 gene is associated with an increased risk of breast cancer. The mis-splicing of the SMN1 gene is associated with spinal muscular atrophy, a neurodegenerative disorder that affects infants and young children.
Methods for Studying Alternative RNA Splicing: Which Of The Following Processes Correctly Describes Alternative Rna Splicing
There are a variety of experimental techniques that can be used to study alternative RNA splicing. These techniques include:
- RT-PCR:RT-PCR can be used to amplify specific RNA transcripts, including alternative splice variants. This technique can be used to determine the expression levels of different splice variants and to identify the factors that influence splice site selection.
- Microarrays:Microarrays can be used to measure the expression levels of thousands of genes simultaneously. This technique can be used to identify genes that are differentially spliced in different cell types or under different conditions.
- RNA-Seq:RNA-Seq is a high-throughput sequencing technique that can be used to identify and quantify all of the RNA transcripts in a cell. This technique can be used to identify alternative splice variants and to study the regulation of alternative RNA splicing.
There are also a variety of bioinformatics tools available for analyzing alternative RNA splicing data. These tools can be used to identify alternative splice variants, to determine the expression levels of different splice variants, and to identify the factors that influence splice site selection.
Expert Answers
What is the role of spliceosomes in alternative RNA splicing?
Spliceosomes are macromolecular complexes that recognize and assemble at specific RNA sequences called splice sites. They catalyze the removal of introns and the joining of exons during RNA splicing. In alternative RNA splicing, different spliceosomes can recognize different splice sites, leading to the production of different mature RNA molecules.
How does alternative RNA splicing contribute to protein diversity?
Alternative RNA splicing can generate multiple protein isoforms from a single gene by combining different exons in different ways. This process allows for the production of proteins with distinct functions, subcellular localization, or regulatory properties, contributing to the vast diversity of proteins in living organisms.
What are some examples of diseases associated with defects in alternative RNA splicing?
Defects in alternative RNA splicing have been linked to various human diseases, including cancer, neurodegenerative disorders, and genetic syndromes. These defects can disrupt the production of functional proteins, leading to cellular dysfunction and disease.